FireBrowse Homepage and BROAD GDAC.
Analysis with FIREHOSE plus TCGA UCSC Xena.
In Brief
-
FIREHOSE Usage.
-
FIREHOSE plus TCGA UCSC Xena.
FIREHOSE Usage
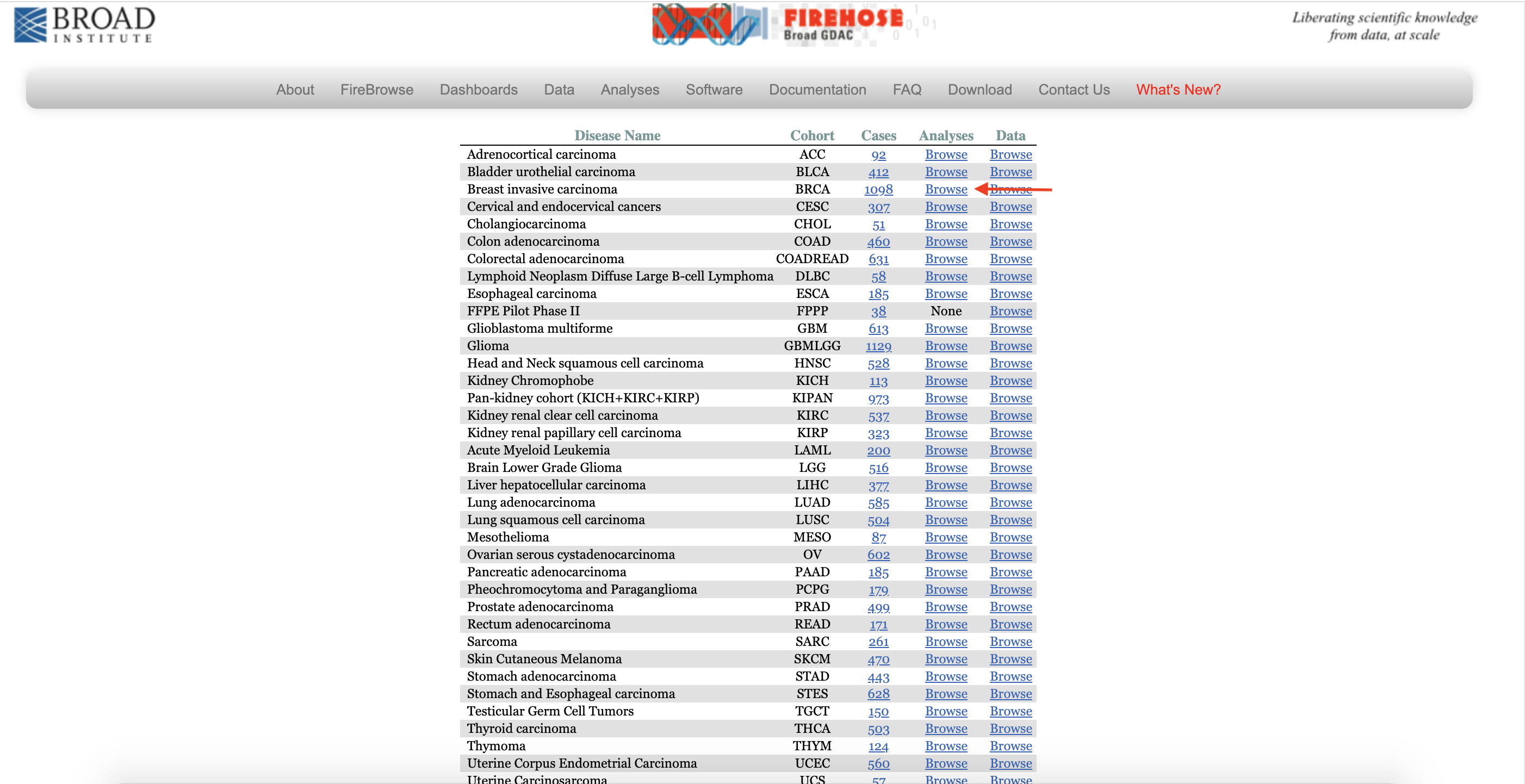
1. Interested Disease from FIREHOSE
-
1.1 Choose Breast invasive carcinoma.
-
1.2 Click on Browse of Analyses from Diseases list.
2. Get Reports of Analysis
-
Click on BRCA of
TCGA data version 2016_01_28 for BRCAto get the reports.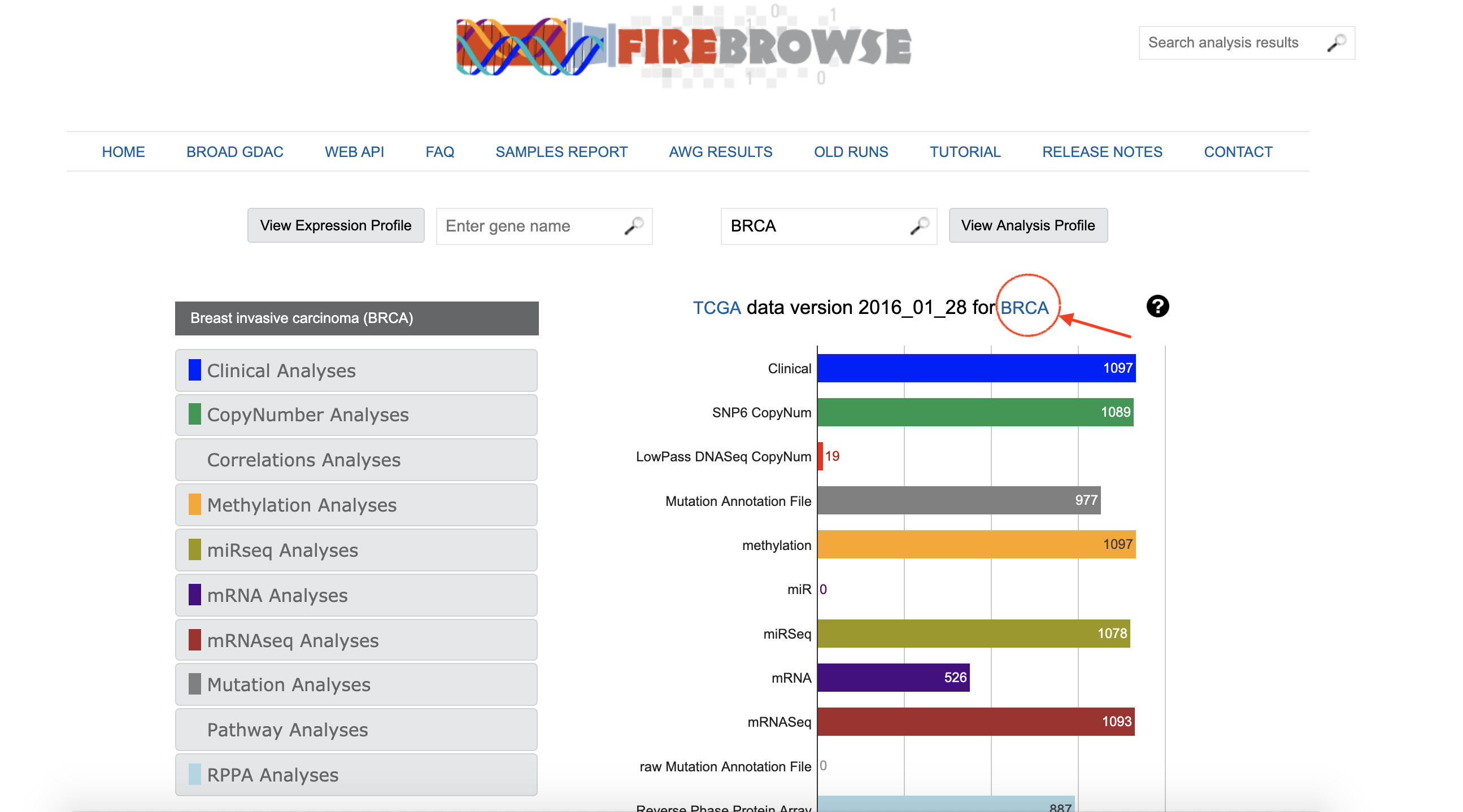
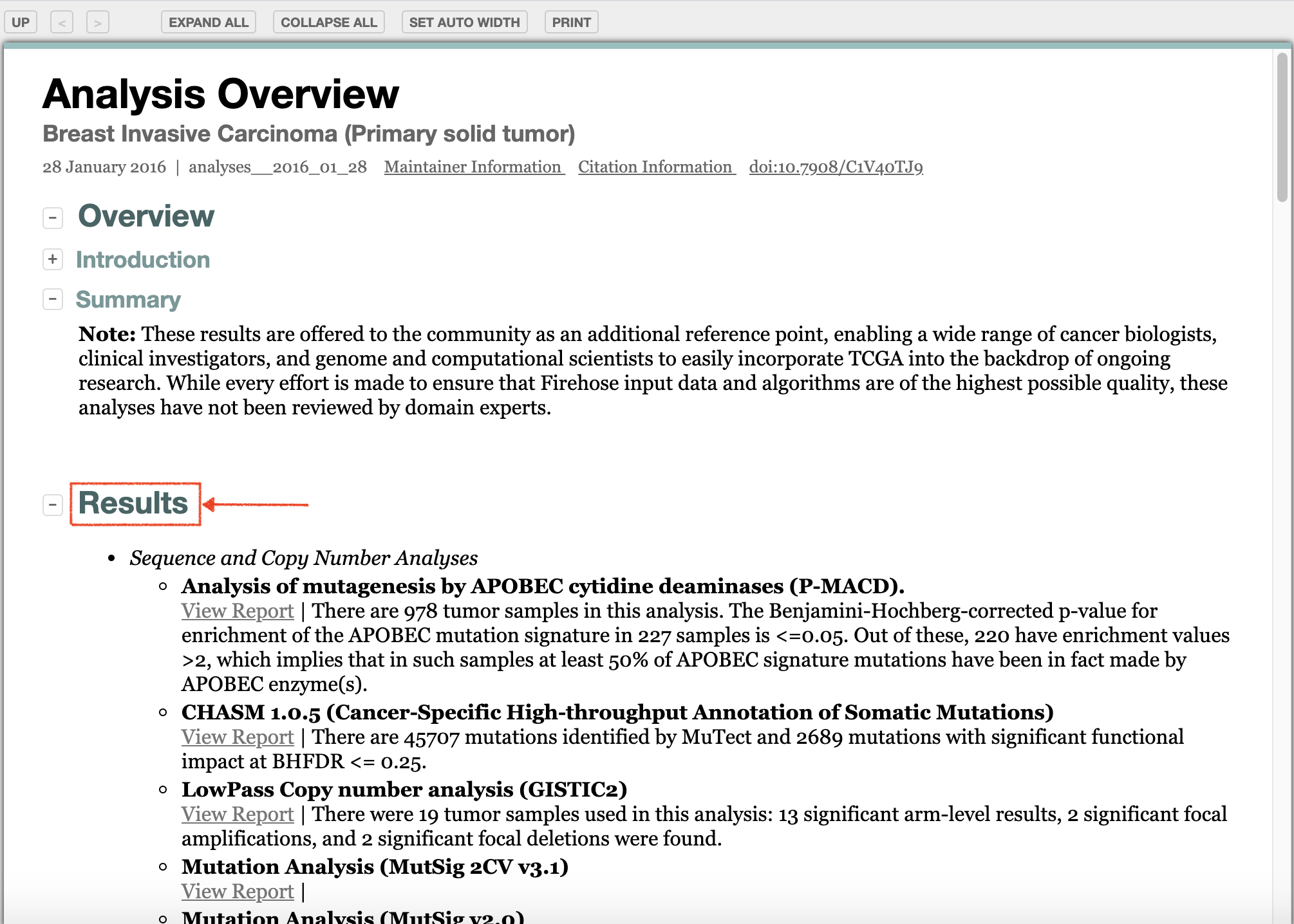
3. Result Analysis
-
In the Results section, choose interested index.
FIREHOSE plus TCGA UCSC Xena
Example 1
-
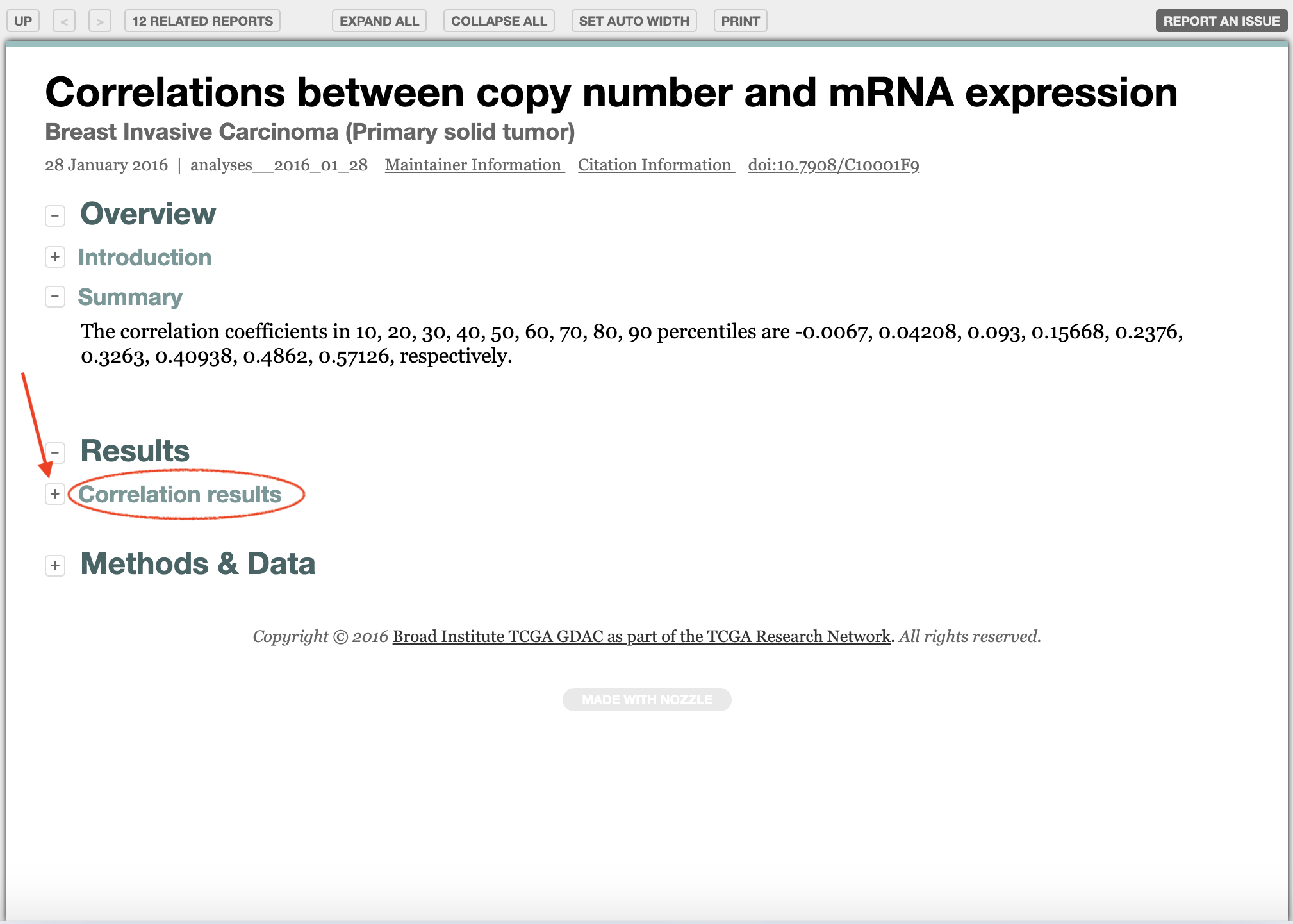
Click on
View Reportof Correlations between copy number and mRNAseq expression.
-
Click on Correlation results.
1. Interested gene, ERBB2
-
Focus on
ERBB2gene, which is very famous gene.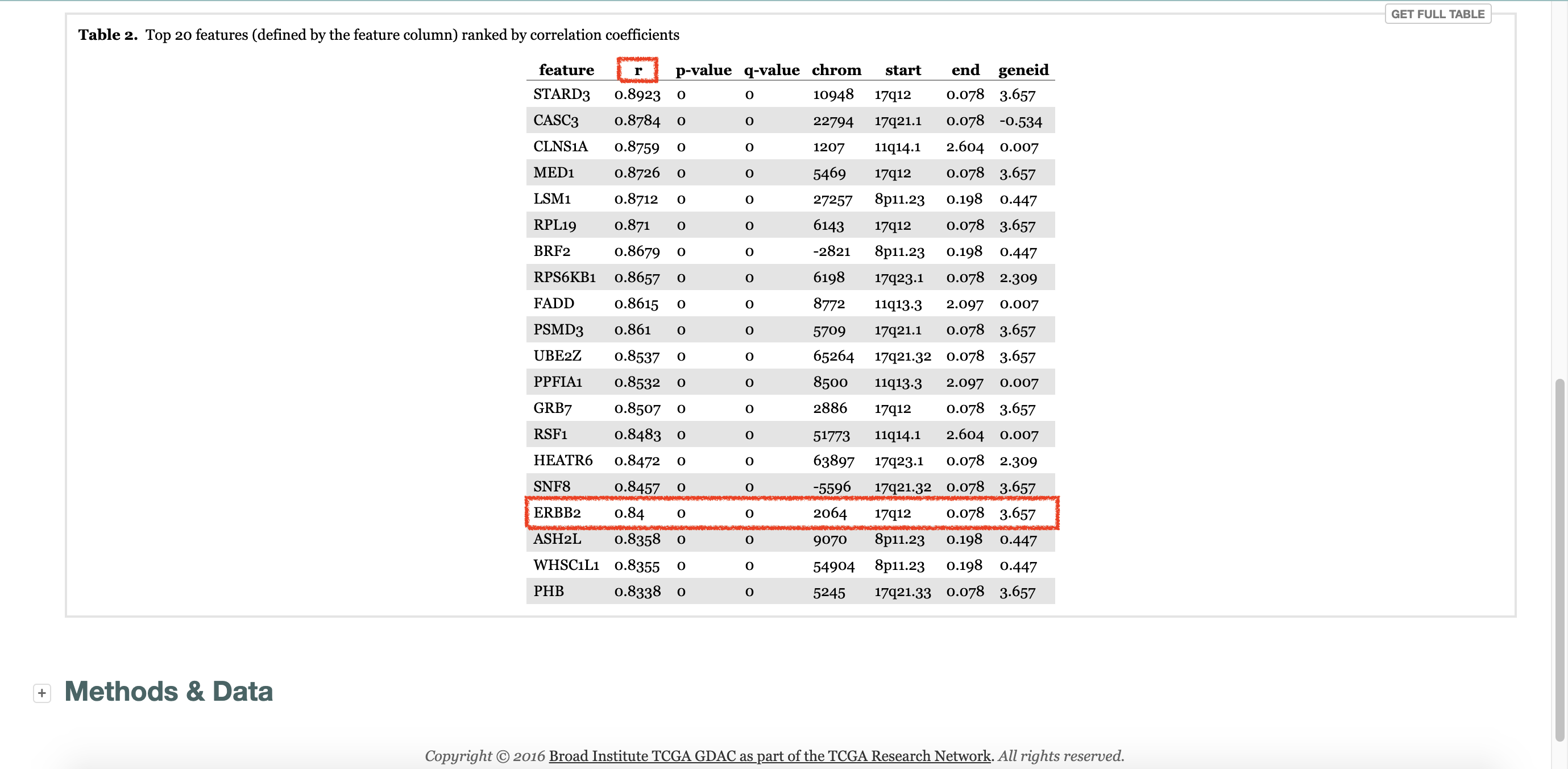
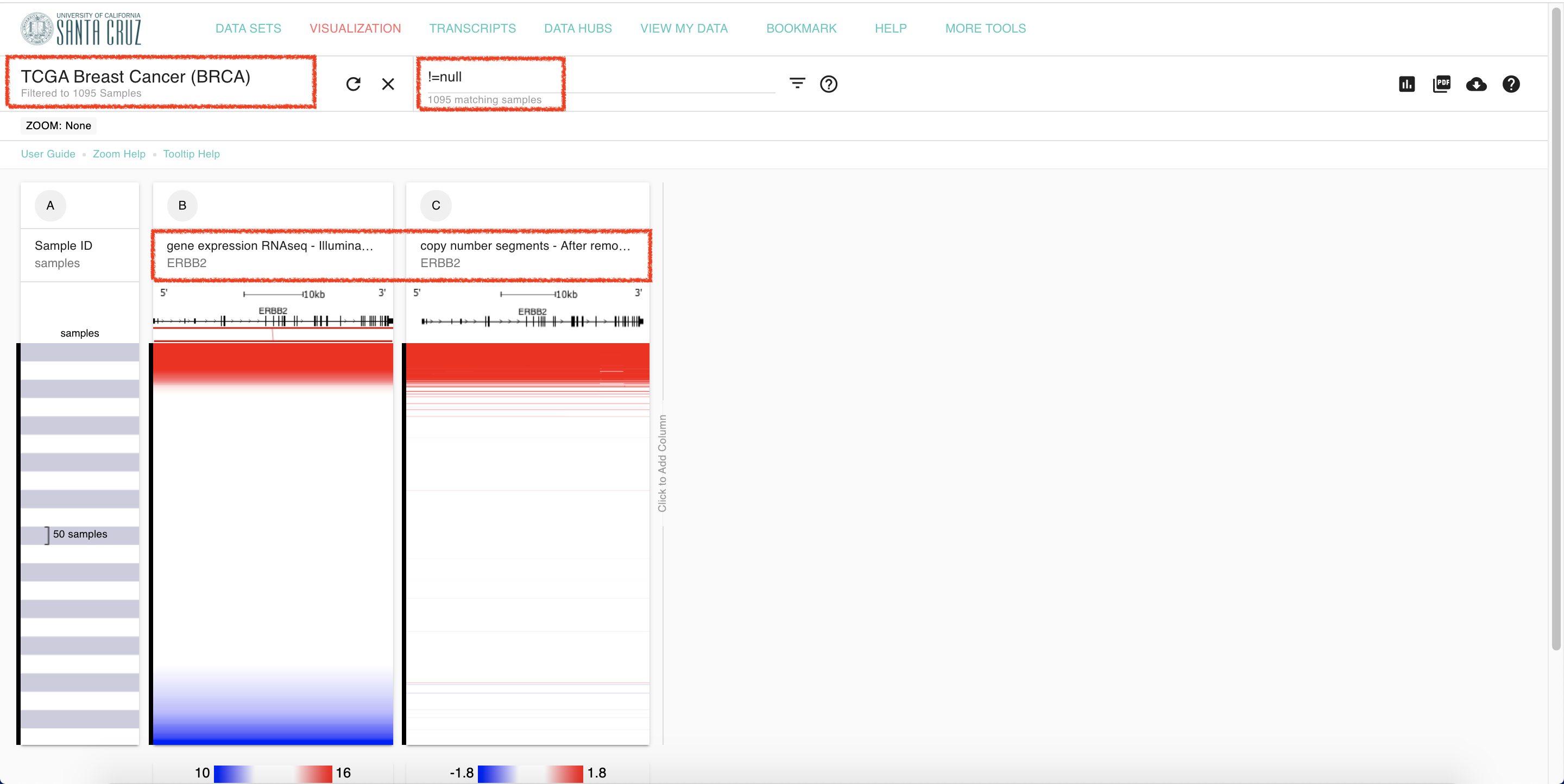
2. Settings for ERBB2
-
Set up ERBB2 gene on UCSC Xena according to firehose.
3. Figure 1
-
As show in figure1,
r = 0.8617 (p = 0.000), which is almost the same as that of firehose Database.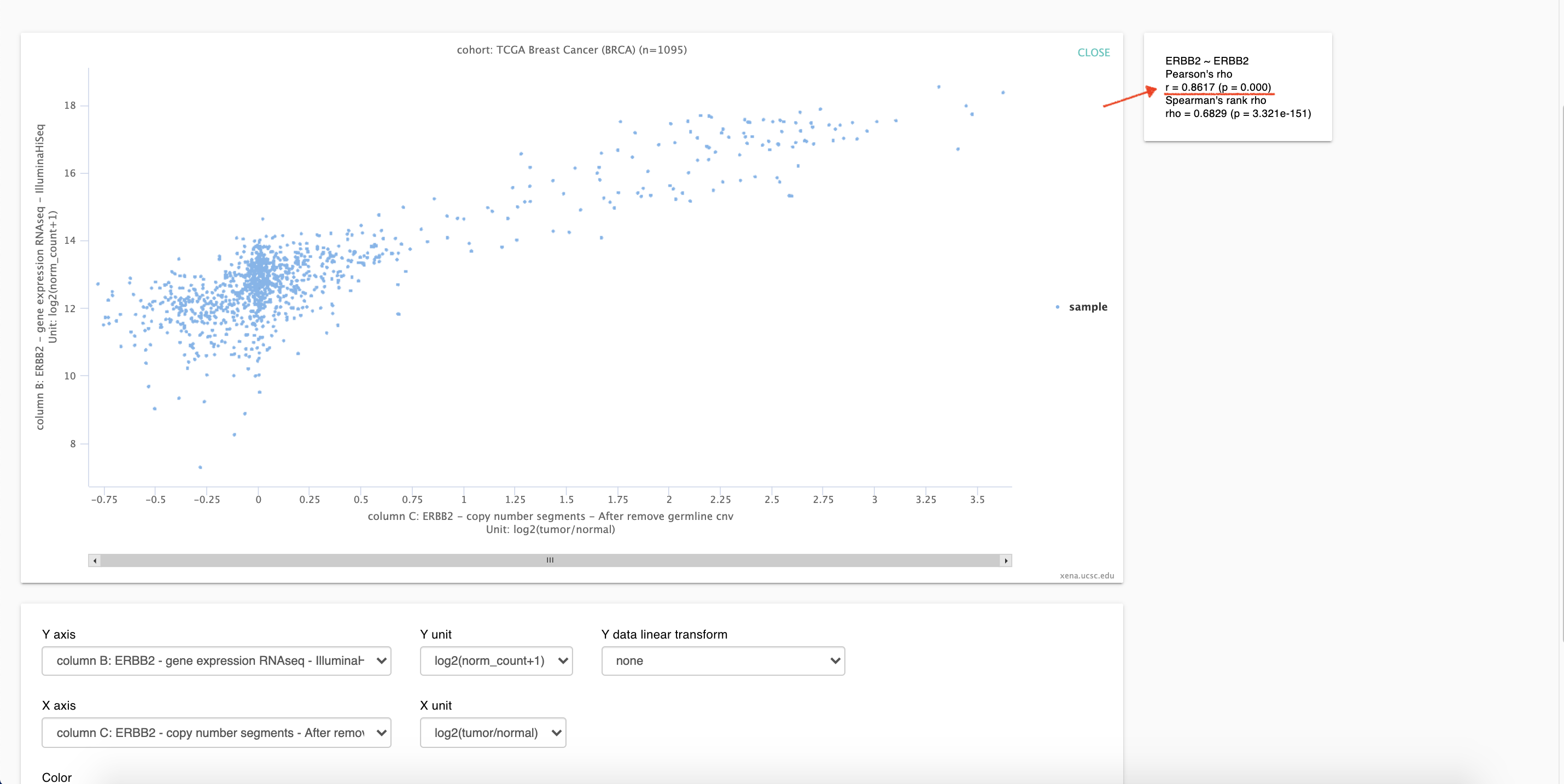
Example 2
-
Click on
View Reportof Correlation between mRNA expression and DNA methylation.
-
Click on Negative Correlation between Methylation and Gene Expression.

1. Interested gene, MKRN3
-
Focus on
MKRN3gene, which ranked No.1 in the list.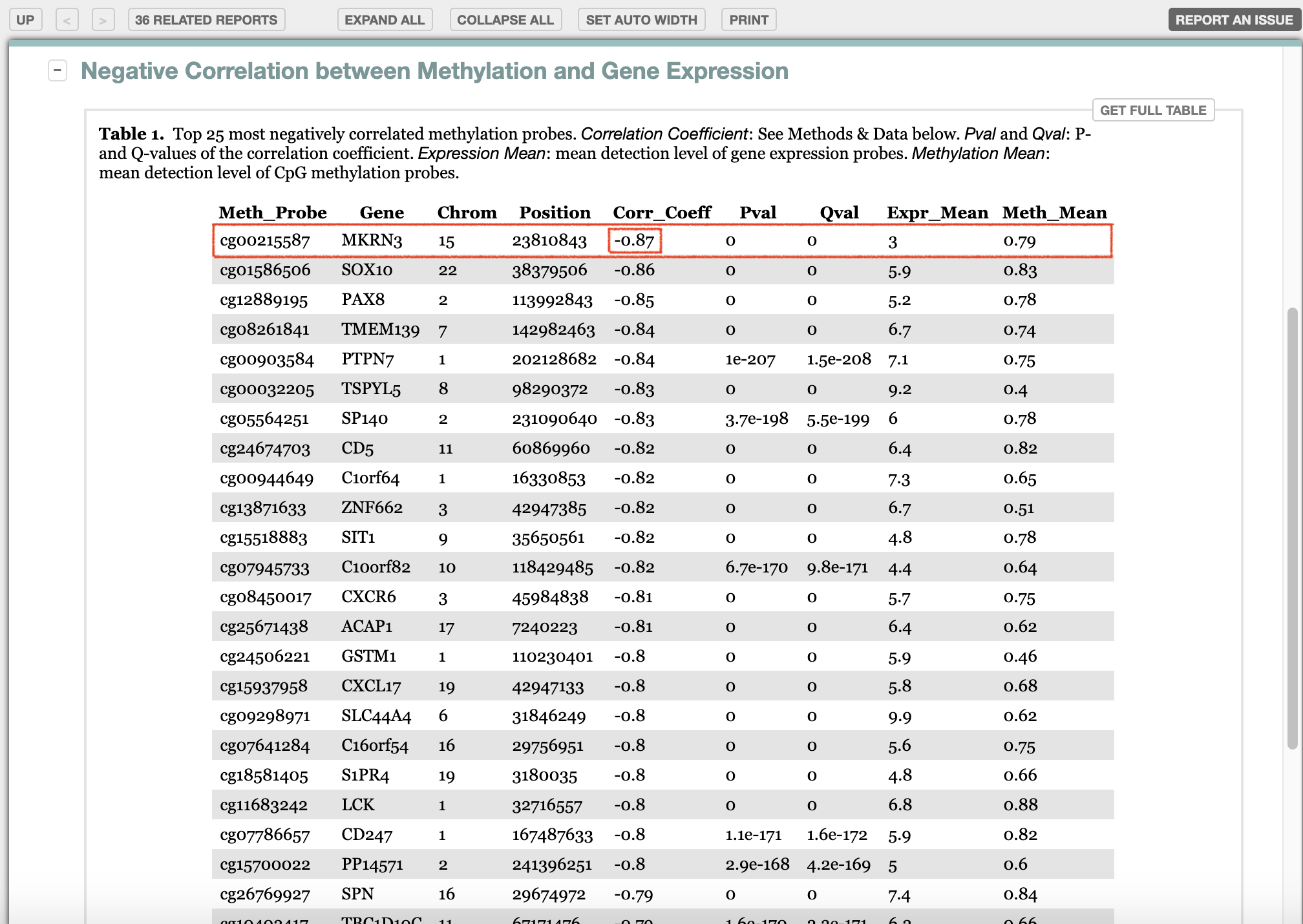
2. Settings for MKRN3
-
Set up MKRN3 gene on UCSC Xena according to firehose.
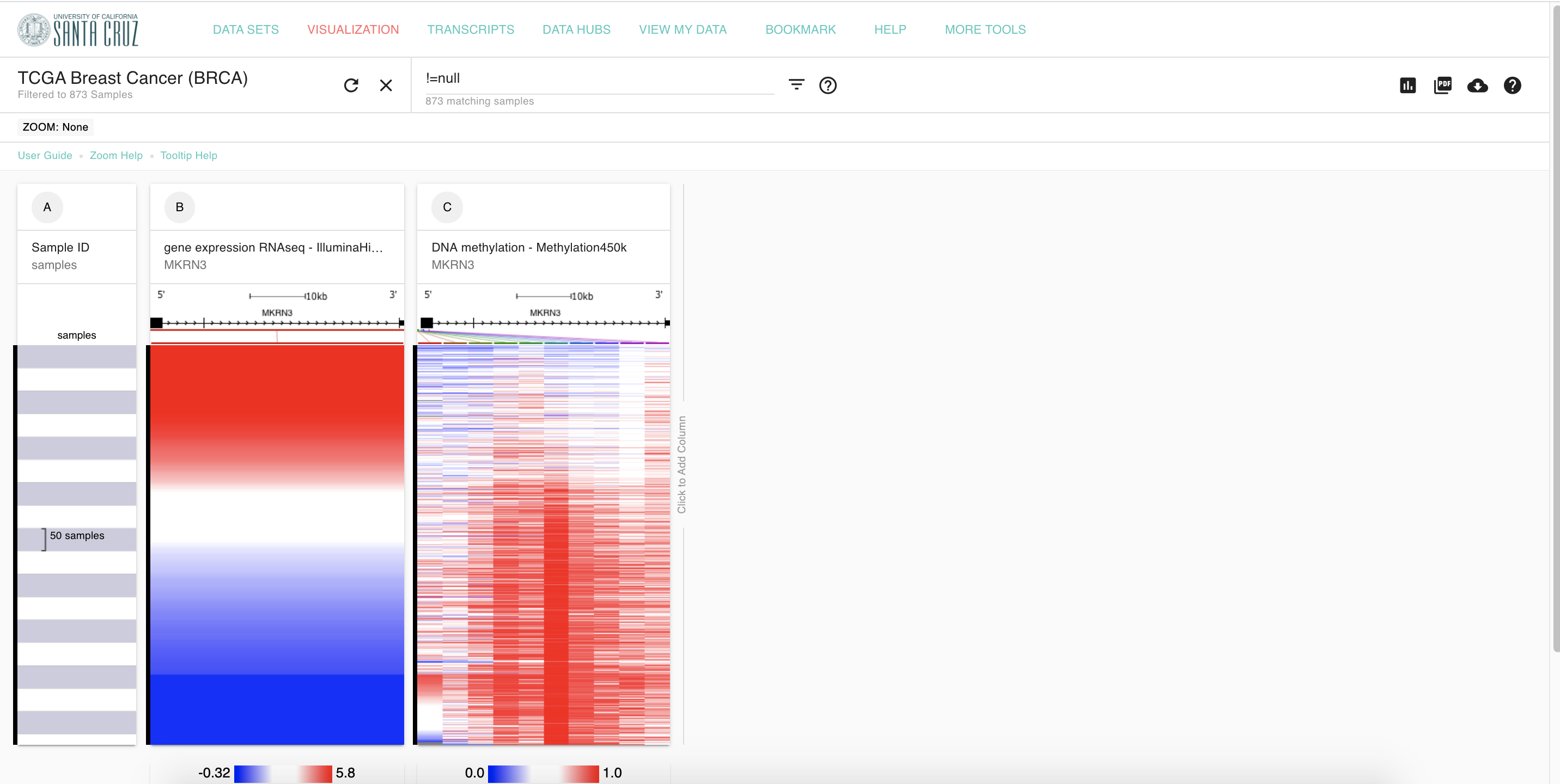
3. Figure 2
-
As show in figure2,
r = -0.8392 (p = 1.538e-232), which is close to that of firehose Database.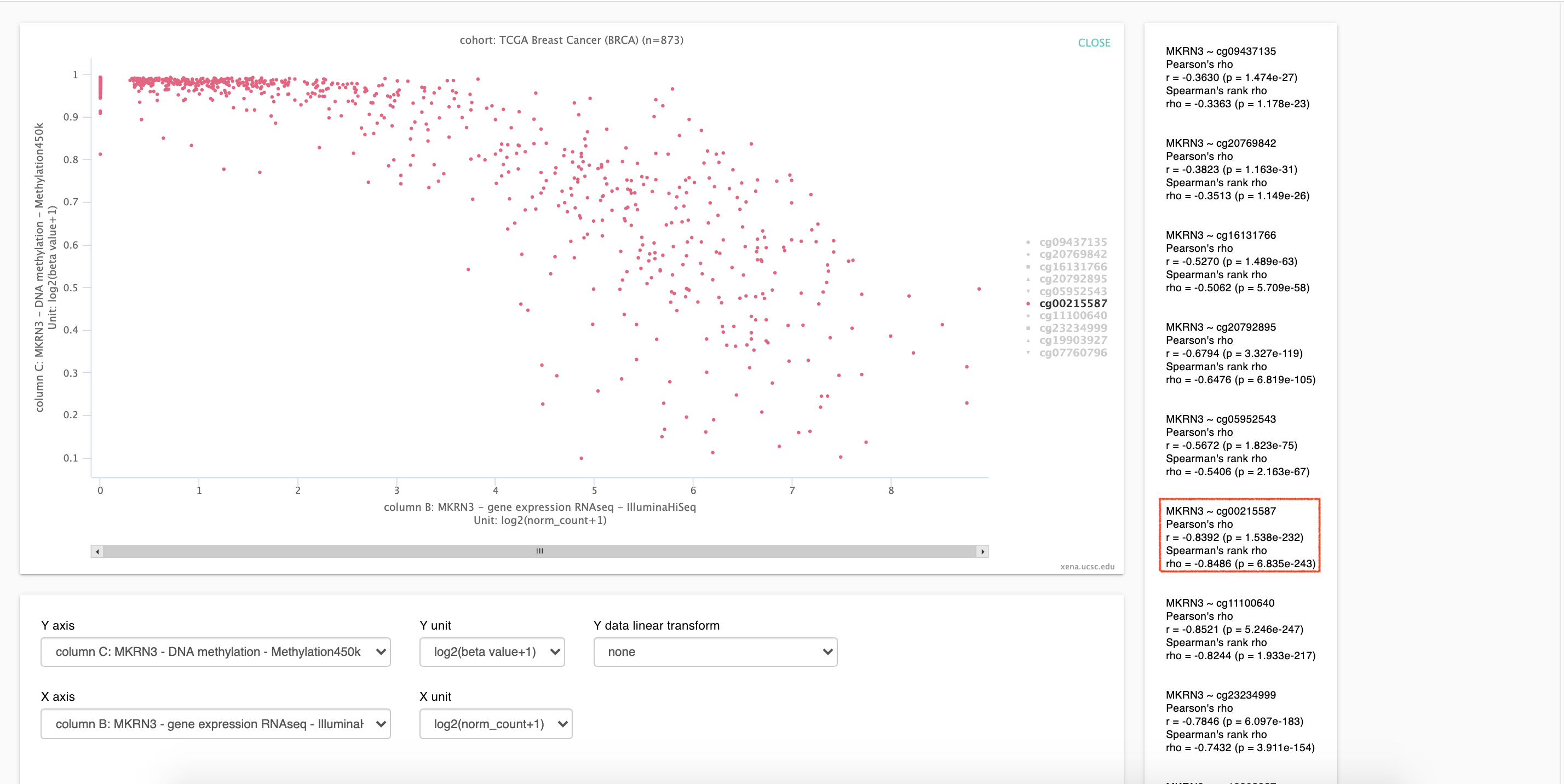
In Summary
-
Different database resources, but similarly conclusion.
-
Start with preliminary conclusion get from bioinformatics analysis, we can go through to verify by performacing related experiments.
-
Start with verified data from experiments, we can furthermore get supported by bioinformatics analysis.