Recent clinical successes of cancer immunotherapy necessitate the investigation of the interaction between malignant cells and the host immune system.
Herein, I summarized five bioinformatics tools for comprehensive analysis of tumor-infiltrating immune cells.
1. TIMER
-
TIMER, A web server for comprehensive analysis of tumor-infiltrating immune cells1.
-
Levels of six tumor-infiltrating immune subsets are pre-calculated for 10,897 tumors from 32 cancer types.
-
6 major analytic modules that allow users to interactively explore the associations between immune infiltrates and a wide-spectrum of factors, including
gene expression,clinical outcomes,somatic mutations, andsomatic copy number alterations.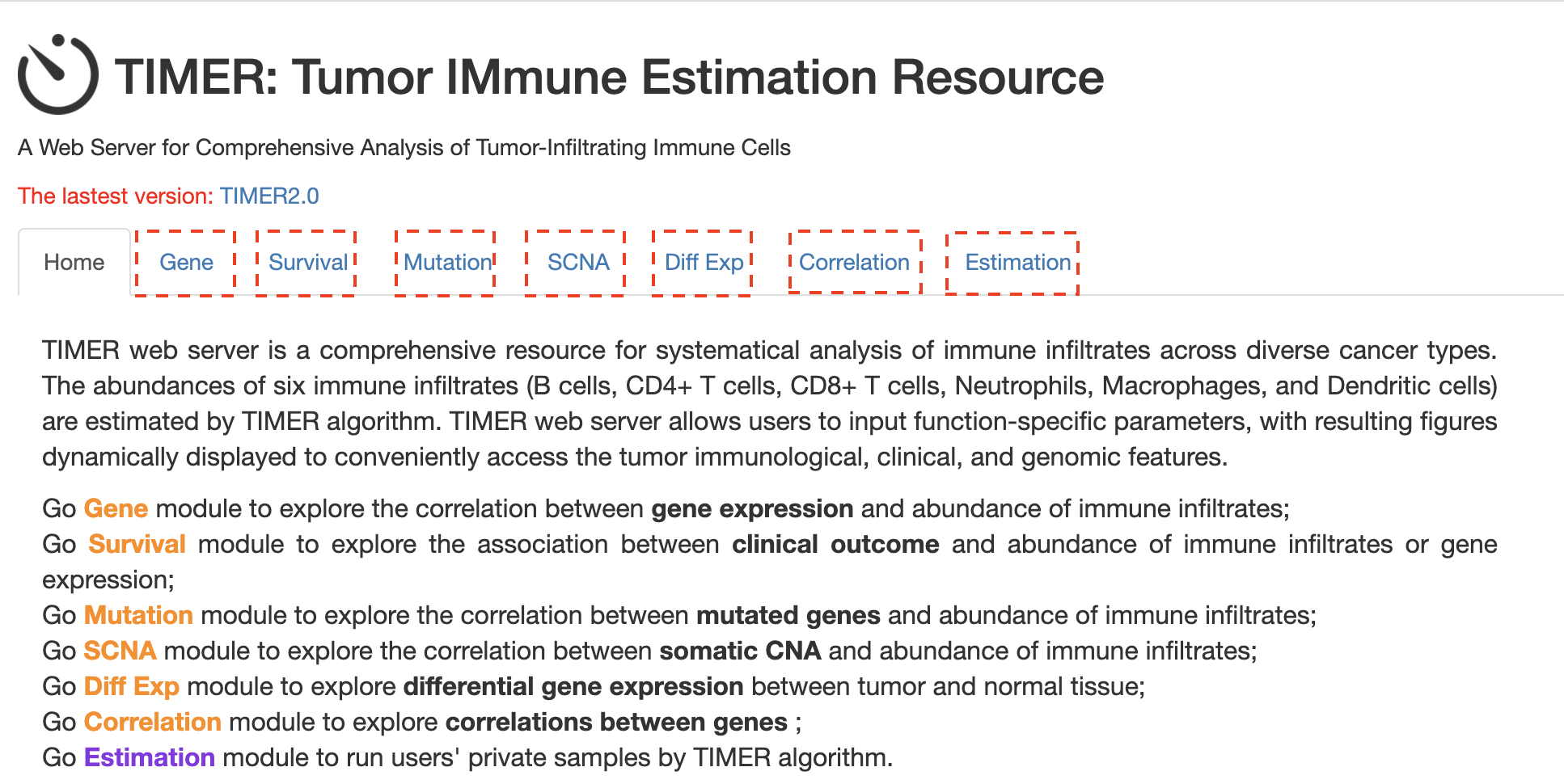
2. The Cancer Immunome Atlas
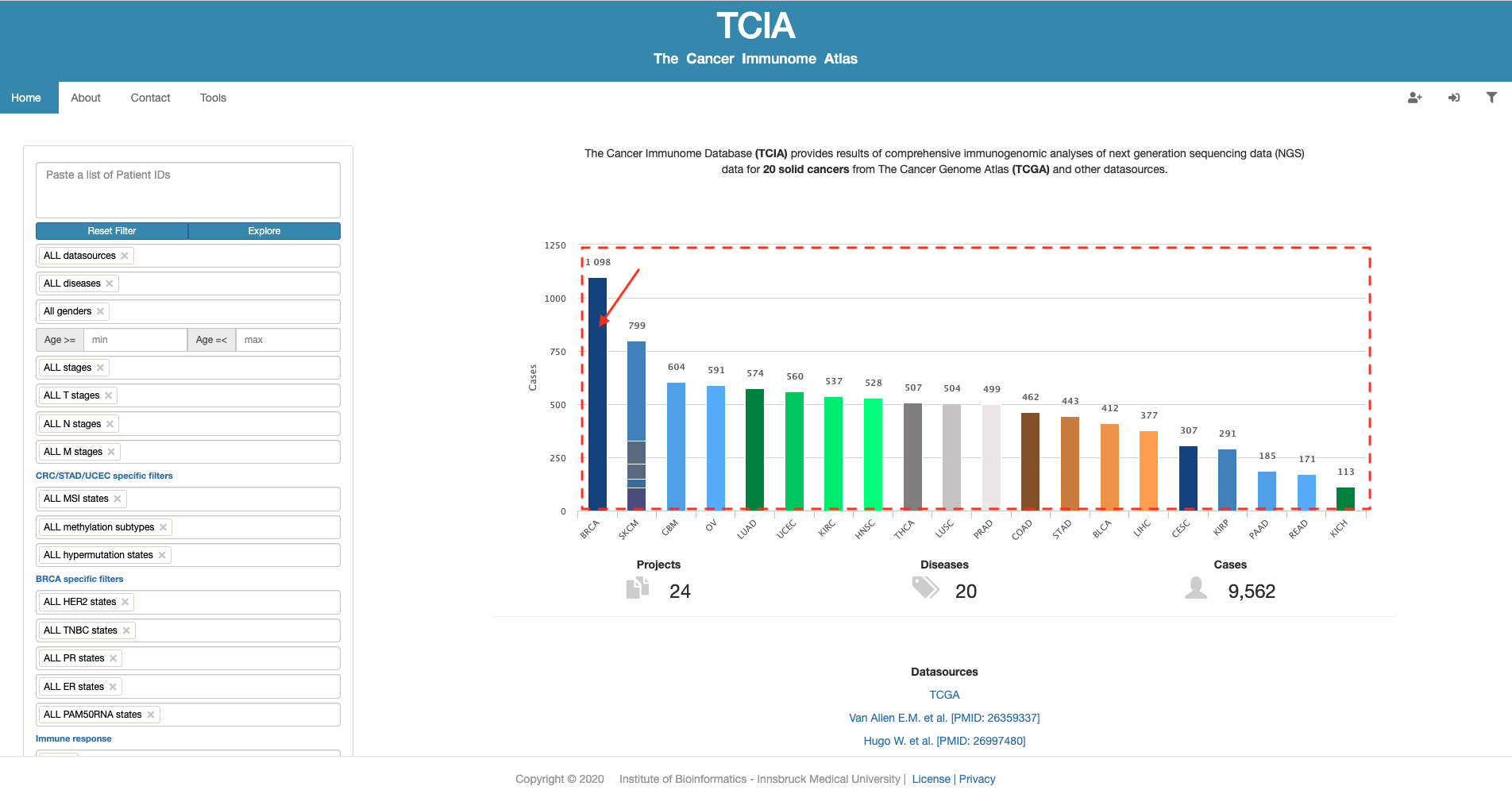
- TCIA, provides results of comprehensive immunogenomic analyses of next generation sequencing data (NGS) data for 20 solid cancers from The Cancer Genome Atlas (TCGA) and other datasources2.
- Cancer genotypes determine tumor immunophenotypes and tumor escape mechanisms
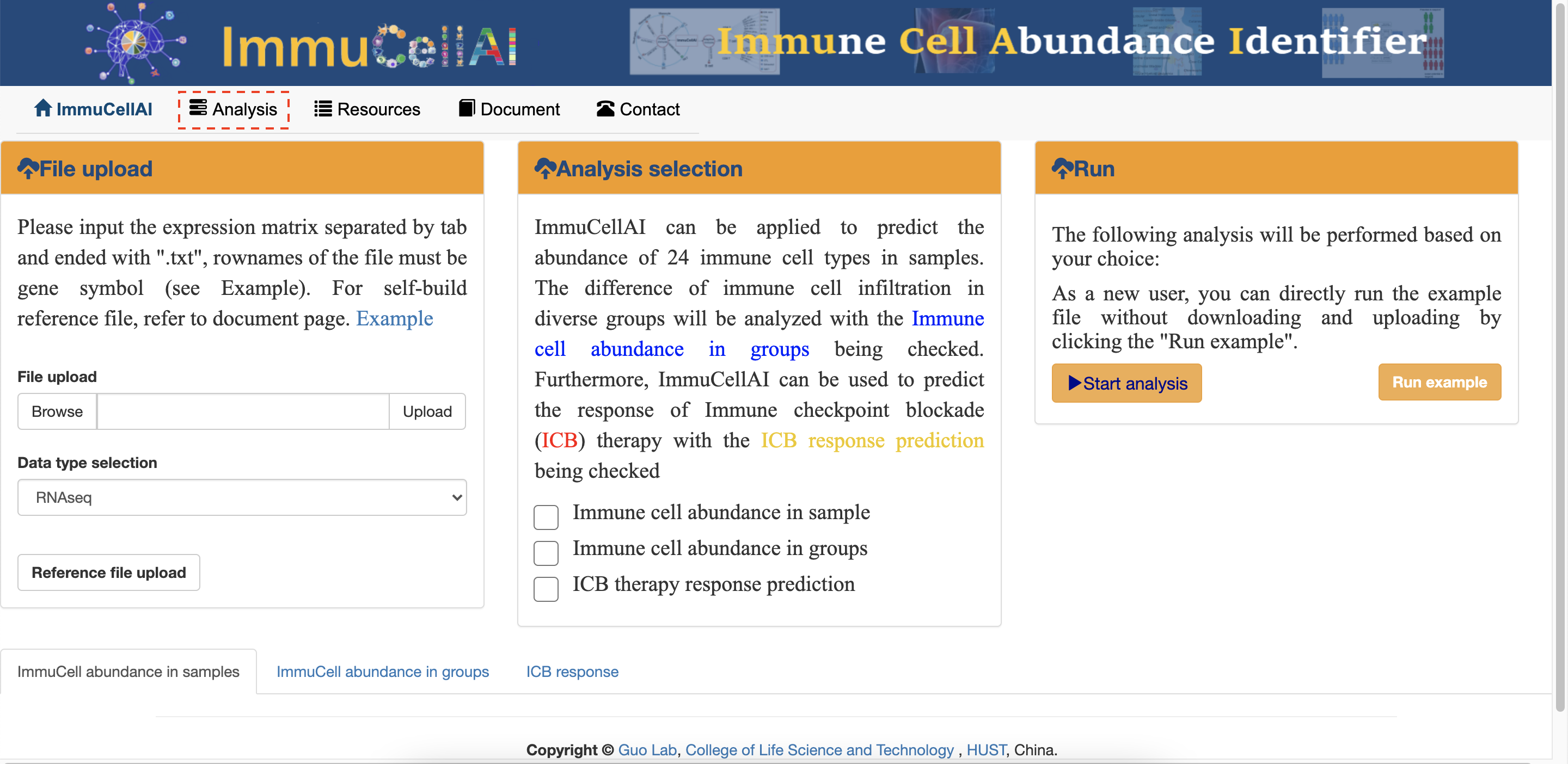
3. ImmuneCellAI
-
ImmuneCellAI, is a tool to estimate the abundance of 24 immune cells from gene expression dataset including RNA-Seq and microarray data, in which the 24 immune cells are comprised of 18 T-cell subtypes and 6 other immune cells: B cell, NK cell, Monocyte cell, Macrophage cell, Neutrophil cell and DC cell3.
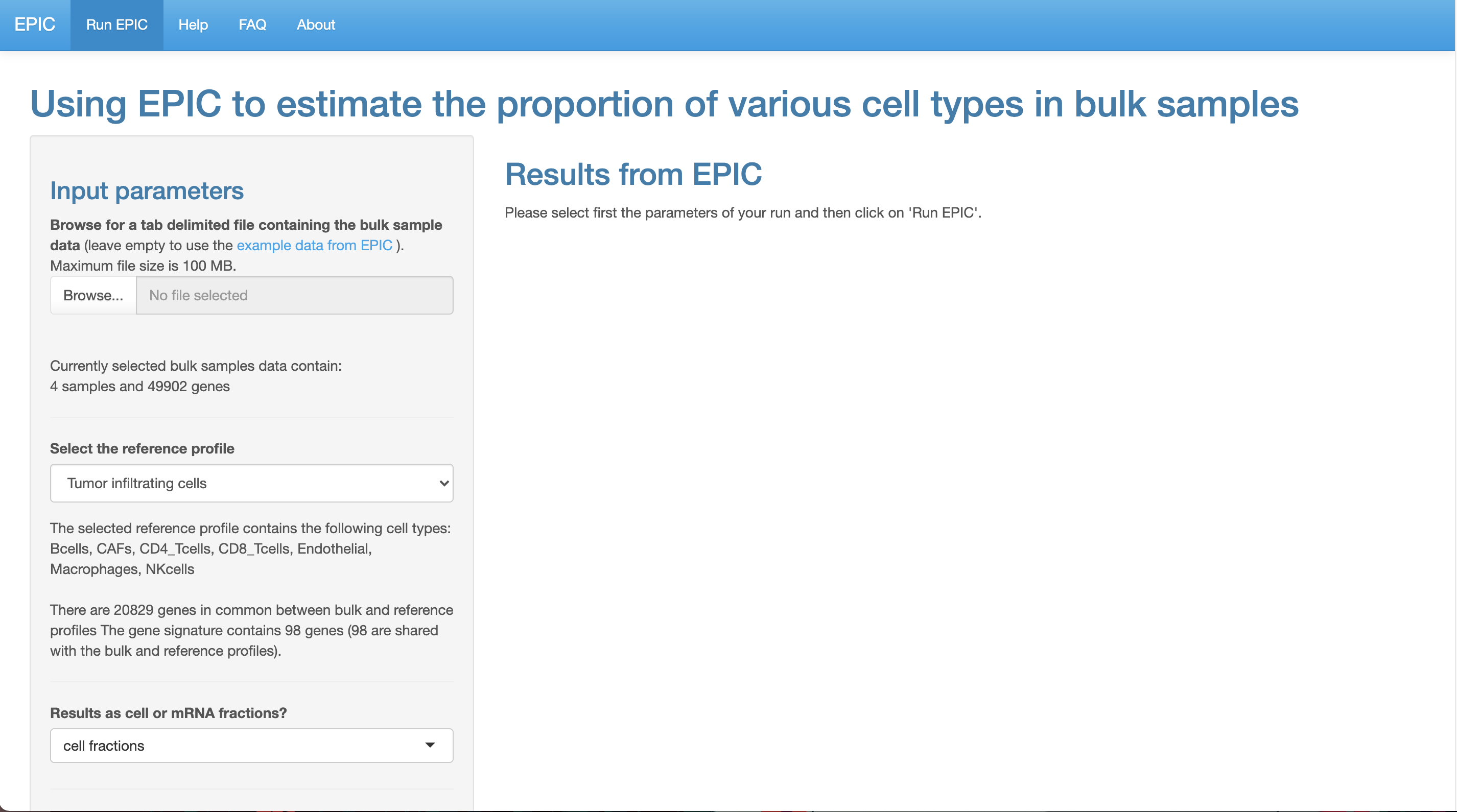
4. EPIC
-
EPIC, is designed to Estimate the Proportion of Immune and Cancer cells from bulk tumor gene expression data4.
5. xCell
-
xCell, is a webtool that performs cell type enrichment analysis from gene expression data for 64 immune and stroma cell types. xCell is a gene signatures-based method learned from thousands of pure cell types from various sources5.

-
xCellView, visualize cell types enrichments.
-
For single-cell RNA-seq, to github and use SingleR.