1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
|
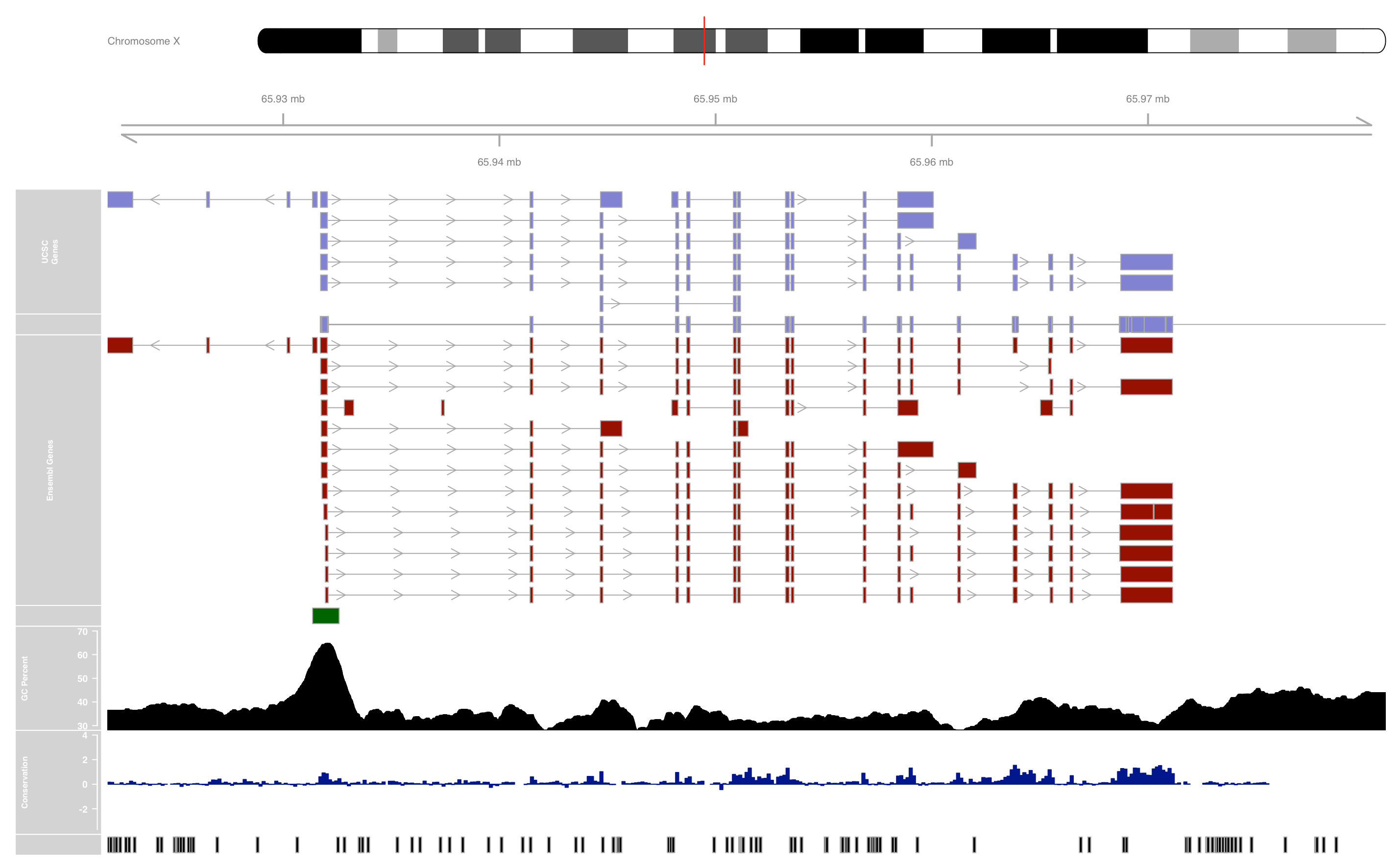
conservation <- UcscTrack(genome = "mm9", chromosome = "chrX",
track = "Conservation",
table = "phyloP30wayPlacental",
from = from, to = to, trackType = "DataTrack",
start = "start", end = "end", data = "score",
type = "hist", window = "auto",
col.histogram = "darkblue",
fill.histogram = "darkblue",
ylim = c(-3.7, 4), name = "Conservation")
gcContent <- UcscTrack(genome = "mm9", chromosome = "chrX",
track = "GC Percent", table = "gc5Base",
from = from, to = to, trackType = "DataTrack",
start = "start", end = "end", data = "score",
type = "hist", window = -1, windowSize = 1500,
fill.histogram = "black", col.histogram = "black",
ylim = c(30, 70), name = "GC Percent")
|